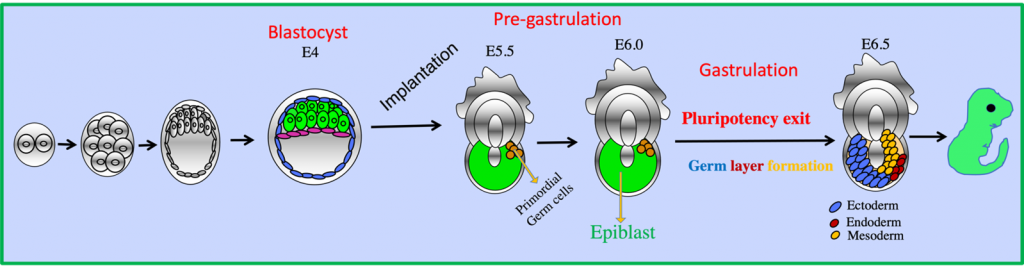
Epigenetic regulation in early embryogenesis, X-chromosome inactivation
Emerging evidence implicates that epigenetics plays a major role in development and disease. Unlike irreversible mutations in DNA, epigenetic modifications are reversible. This inherent plasticity makes epigenetic changes associated with diseases potentially amenable to manipulation via therapeutic intervention. However, much about the mechanistic aspects of epigenetic regulation remains to be understood. Our research strives to further the understanding of mechanism of epigenetic regulation through the study of X-chromosome inactivation, imprinting and stochastic allelic gene expression during early-mammalian development using mouse/human embryos, germ cells and pluripotent stem cells. The long-term goal of these studies is not only to advance our understanding of basic epigenetic mechanisms during development, but also to extend the knowledge from the early development to (a) cancer stem cell biology to better understand cancer pathogenesis and provide therapeutic intervention (b) to develop a clinical strategy to prevent the gender bias issue in babies born through in vitro fertilization (IVF) and enhancement of IVF success rate and (c) improving the quality of human iPSC.